Scientific Achievement
Proposed and cross-validated experimental and computational workflows to analyze structural populations of disordered polypeptoids.
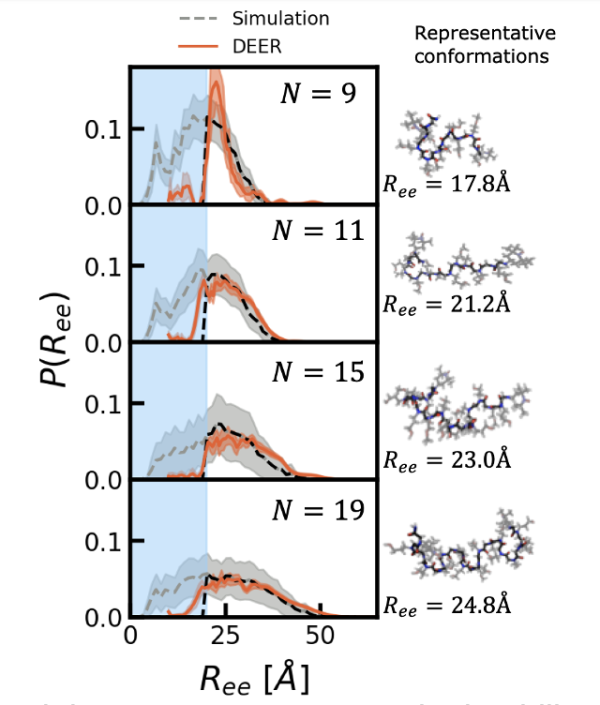
Image: End-to-end distributions, 𝑃(𝑅𝑒𝑒 ), for hydrophilic polypeptoids of varying length show excellent agreement between simulations (dashed black lines) and experiments (orange solid lines). Snapshots on the right show example structures close to the mean end-to-end distance for each length.
Significance and Impact
This combined workflow overcomes challenges in characterizing disordered polypeptoids, enabling studies of sequence-structure-function relationships.
Research Details
- Polypeptoids provide a robust and precisely controllable platform for analyzing the effect of sequence on functional properties, but disordered polypeptoids are poorly characterized.
- An efficient enhanced sampling algorithm overcomes long timescales in polypeptoid simulations.
- Computational and experimental end-to-end distance distributions are in excellent agreement and both suggest excluded volume scaling of polypeptoids.
Notes
Polypeptoids are bio-inspired polymers that can be synthesized sequence-specifically, which makes them an attractive platform for studying the effects of heterogeneous chemistries. However, the structural properties of long, disordered polypeptoids are poorly understood. Most experimental characterization techniques cannot resolve the full distribution of conformations exhibited by disordered molecules. Furthermore, while computational models (forcefields) for polypeptoids have been developed, they have only been tested on short, structured polypeptoids. In this work, we developed an integrated experimental-computational approach offering unprecedented insight into the structural behavior of disordered polypeptoids by measuring the probability distributions of distances between polypeptoid ends using both simulations and Double Electron-Electron Resonance (DEER). The distributions were in excellent agreement (see figure), demonstrating convergence of the experimental and computational techniques. The distributions furthermore showed, by comparison to ideal polymer models, that polypeptoids with polar sidechains adopt conformations similar to well-hydrated polymers (excluded volume scaling). As part of this project, we also developed a simulation workflow that utilizes enhanced sampling techniques to sample over slow backbone conformational changes that are inaccessible by conventional molecular dynamics techniques. This generalizable workflow scales well with length, which makes it unique among approaches typically taken.
Acknowledgements
This work was supported by the Center for Materials for Water and Energy Systems (M-WET), an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences under Award #DE-SC0019272. Use was made of computational facilities purchased with funds from the National Science Foundation (OAC-1925717) and administered by the Center for Scientific Computing (CSC). The CSC is supported by the California NanoSystems Institute and the Materials Research Science and Engineering Center (MRSEC; NSF DMR 1720256) at UC Santa Barbara. S.J., J.I.M., and M.B. acknowledge support by the National Science Foundation Graduate Research Fellowship (DGE 1650114, DGE 1144085). M.B. also recognizes support from the Office of Naval Research (ONR) under Award #N00014-17-1-2047. A.D. acknowledges support by the National Defense Science & Engineering Graduate (NDSEG) Fellowship Program. The authors gratefully acknowledge access to the WavPDS and SVDReconstruction software through the National Biomedical Center for Advanced ESR Technology (ACERT). ACERT is hosted by the Cornell Center for Advanced Computing and supported by the National Institute of General Medical Sciences. Conversations with M. Srivastava regarding the use of WavPDS and SVDReconstruction were particularly helpful. The authors acknowledge the use of MRL Shared Experimental Facilities, which are supported by the MRSEC Program of the NSF under Award No. DMR 1720256, a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org). The authors also acknowledge helpful discussions regarding DEER acquisition and analysis with T. Keller and K. Tsay and conversations with R. Zuckermann regarding polypeptoid synthesis. The authors thank A. Prakash and J. Pfaendtner for providing modified AMBER99SB-ILDN forcefield files for peptoids and helpful discussions concerning their implementation and R. Jain and E. Santiso for helpful discussions concerning the implementation of the modified CGenFF forcefield.
Related Publication
Sally Jiao, Audra DeStefano, Jacob I. Monroe, Mikayla Barry, Nicholas Sherck, Thomas Casey, Rachel A. Segalman, Songi Han, and M. Scott Shell, "Quantifying Polypeptoid Conformational Landscapes through Integrated Experiment and Simulation," Macromolecules, 54(11), 5011-5021, 2021 May 25, https://doi.org/10.1021/acs.macromol.1c00550.

