Scientific Achievement
Proposed and cross-validated experimental and computational workflows to analyze structural populations of disordered polypeptoids.
Significance and Impact
This combined workflow overcomes challenges in characterizing disordered polypeptoids, enabling studies of sequence-structure-function relationships.
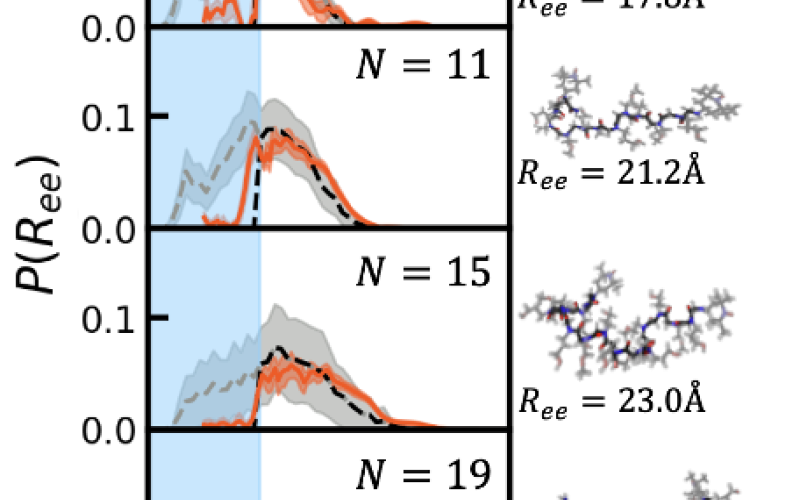
End-to-End Distributions
End-to-end distributions, P(R_ee), for hydrophilic polypeptoids of varying length show excellent agreement between simulations (dashed black lines) and experiments (orange solid lines). Snapshots on the right show example structures close to the mean end-to-end distance for each length.
Research Details
- Polypeptoids provide a robust and precisely controllable platform for analyzing the effect of sequence on functional properties, but disordered polypeptoids are poorly characterized.
- An efficient enhanced sampling algorithm overcomes long timescales in polypeptoid simulations.
- Computational and experimental end-to-end distance distributions are in excellent agreement and both suggest excluded volume scaling of polypeptoids.
We combine experiment and simulation to develop a powerful, validated approach for characterizing the conformational landscapes of disordered polypeptoids. Polypeptoids have become an important class of polymers, capable of precisely defined sequences while remaining gram-synthesizable – properties that have driven a rapidly expanding set of applications, including antifoulants, therapeutics, sensing, and directed self-assembly. The characterization of polypeptoid structure provides critical molecular insight into sequence-structure-function relationships. Structurally disordered polypeptoids require new approaches to interrogate their wide range of conformations in solution. Here, we measure full end-to-end distance distributions, instead of configurational averages, using Double Electron-Electron Resonance (DEER) spectroscopy and enhanced-sampling molecular modeling. We demonstrate excellent agreement between the experiments and simulations for a set of model hydrophilic polypeptoids. Moreover, we illustrate the utility of this combined experiment-simulation approach in probing structure-function relationships by characterizing the basic polymer physics of this polypeptoid series, demonstrating that the polypeptoids probed here exhibit excluded volume behavior.
Sally Jiao, Audra DeStefano, Jacob Monroe, Mikayla Barry, Nicholas Sherck, Thomas Casey, Rachel Segalman, Songi Han, M. Scott Shell, Macromolecules (2021)
Work was performed at UC Santa Barbara.

